Lists: ArrayLists and LinkedLists
ArrayLists
The fact that the length of arrays cannot be modified is a pretty big limitation. What if we don’t know how many elements will have at the beginning? What if we want the length of our list to change in response to something that dynamically happens in our program? For these reasons, we have ArrayLists, which build on our arrays by being able to change their size.
At this point, it is useful to talk about the difference between the size of a list versus the capacity of a list. The size of a list is the number of elements that it currently holds, while the capacity of a list is the number of elements that it could potentially hold. Other than that, an ArrayList is functionally the same as an array that we discussed above. Here are some of the commonly used methods that you can use to manipulate ArrayLists:
add(E element)adds the element of typeEto the end of the list.add(int index, E element)adds the element of typeEat the specified position in the list.clear()removes all elements from the list and resets the size of theArrayListto zero.contains(Object o)returnstrue(aboolean) if the list contains the specifiedObject.ensureCapacity(int minCapacity)increases the capacity of theArrayListto ensure that it can hold at leastminCapacityelements.get(int index)returns the element at the specified position in this list.isEmpty()returnstrue(aboolean) if the list contains no elements (has size zero).remove(int index)removes the element at the specified position in the list.remove(Object o)removes the first occurrence of the specified element from this list, if it is present. If you happen to be working with anArrayListofints, the first definition of theremove()method takes priority.size()returns the number of elements in this list.toArray()converts theArrayListinto an array.set(int index, E element)replaces the element at the specified position in this list with the specified input element.
Other methods can be found in the official Java documentation.
LinkedLists
Last time, we talked about the structure of a list, which we defined to be an ordered array of elements. Arrays had the limitation of having an immutable size and capacity. If we had an array that could hold a maximum of 10 elements and want to add an 11th element, we would have to create an entirely new array with a larger capacity. ArrayLists were able to overcome this limitation by featuring a mutable size and a capacity that could be expanded “under the hood.”
Remember that both arrays and ArrayLists featured contiguous elements. This means that the (i)th and (i+1)th elements are right next to each other in the computer’s memory. For example, let’s say we had an ArrayList that looked something like
import java.util.ArrayList;
public class Example {
public static void main(String[] args) {
ArrayList<Integer> data = new ArrayList<>();
data.add(6);
data.add(-2);
data.add(7);
data.add(0);
data.add(4);
}
}
If we were to represent data conceptually, it might look something like this:
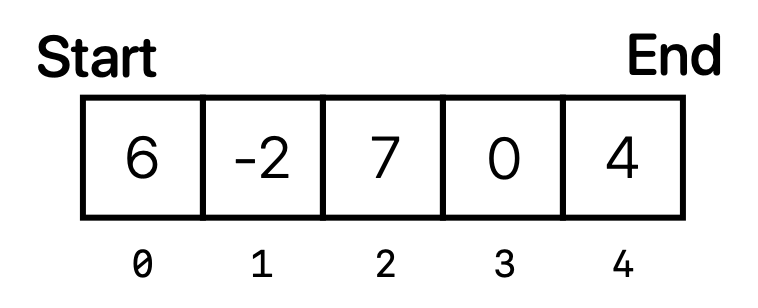
Imagine if we wanted to remove the element data.get(2), which is equal to 7 according to our code. Because elements in arrays and ArrayLists must be contiguous in the computer’s memory, once we call data.remove(2) to remove the second element not only do we have to remove the element 7, but we also have to move the element 0 (the third element) one position to the left, and then the element 4 (currently the fourth element) also one position to the left. This essentially involves copying and moving two elements around. For this simple example, this isn’t too big of an issue. However, what if our data set was much larger? What if data contained 10 million elements, and we wanted to remove the 0th element? Or if we wanted to add an element at the very beginning of the ArrayList? All of a sudden, these processes will start to become very slow because they involve copying and moving so many different elements.
Fortunately for us, LinkedLists provide a better, more time-efficient alternative. LinkedLists are another implementation of the list structure that we talked about previously. The allow for us to store data elements of the same type in an ordered fashion, and from the outside, will look and act exactly like any other type of list. However, LinkedLists have the one caveat where list elements don’t have to be contiguous. Rather, list elements are now called nodes that store a pointer to the next element in the list in addition to the data entry. For now, we can think of a pointer as an arrow that points to the next element. To create a LinkedList, our Java code might look something like this:
import java.util.LinkedList;
public class Example {
public static void main(String[] args) {
LinkedList<Integer> data = new LinkedList<>();
data.add(6);
data.add(-2);
data.add(7);
data.add(0);
data.add(4);
}
}
If we were to represent this version of data conceptually, it might look something like this:

Here, everything looks the same, but we now also have the pointers (denoted as arrows in the diagram) that point to the next element in the list. Imagine if we wanted to remove the element data.get(2), which is equal to 7 according to our code. Calling data.remove(2) to remove the second element only requires two operations: removing the pointer from the 7 to the 0, and “rerouting” the pointer from the 2 to the 0. Therefore, after calling data.remove(2), our LinkedList might look something like this:
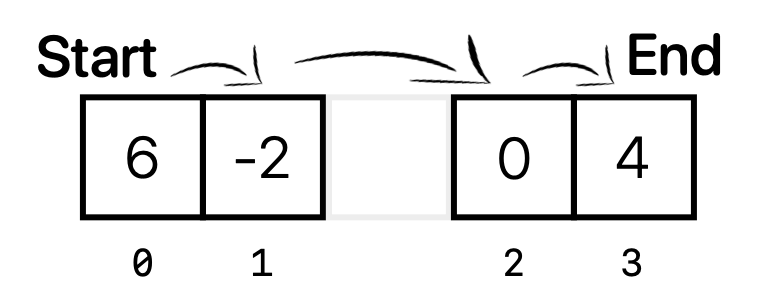
The benefit of a LinkedList is that all of the data elements stay in place! This means that we don’t have to move anything around. All we need to do is update which direction the pointer is pointing. As another example, say we want to add the element 10 to the beginning of this list. After calling data.add(0, 10), data would look like

Again, nothing had to move! However, note that one limitation that arises with LinkedLists is that if we’re trying to look for elements or access elements of the list, we always have to look through all of the elements before our desired element of interest in order to get the pointers to know how to traverse the list correctly. This is because the list nodes are not necessarily contiguous anymore. As a result, if your element is at the end of the list, you’ll have to traverse through all of the elements in the list before you can access it.
As we’ve seen, LinkedLists are another great way to represent lists in Java. In general, if you know that your program will be adding and removing and moving elements around a lot, it might be more advantageous to use LinkedLists. If you know that you’ll be reading and writing elements a lot, it might be more advantageous to use ArrayLists. If you don’t quite know what you’ll need to be doing with your list, it might be worthwhile to try out both and see which one is faster!
Exercises
Problem 1
DNA is composed of four different types of “building blocks,” which we’ll refer to as A, G, C, and T. A valid DNA sequence will be composed of these four types of characters, which can be either uppercase or lower case. For example AcTggCGa is a valid 8-character DNA sequence. Fill in the following methods for our class representation of DNA below:
public class DNA {
// TODO: Include any relevant private fields here.
/*
* Constructor takes in a String sequence input and sets the
* appropriate vales for any of your relevant fields. Make sure to
* confirm that the input sequence is a valid input sequence: that
* is, it only contains A, G, C, T, and/or their lower-case
* equivalents.
*/
public DNA(String sequence) {
// TODO: Do anything you need to do here. Make sure to store
// the input DNA sequence as a field so we can use it in other
// methods below.
}
/*
* This function should return the number of characters in the DNA
* sequence.
*/
public int length() {
// TODO
return -1;
}
/*
* Sometimes, biologists are interested in a DNA sequence's "GC
* content", which is defined to be the percentage of characters
* in the DNA sequence that are G, g, C, or c. Calculate and
* return the GC-content of the DNA sequence.
*/
public double getGCcontent() {
// TODO: returns the percentage of characters in the DNA sequence
// that are G, g, C, or c.
return -1;
}
/*
* This function should compare the original DNA sequence from the
* constructor with another DNA sequence class instance and return
* if they are equal. Two DNA sequences are equal if they contain
* the same sequence of characters (aside from upper and lower case
* differences).
*/
public boolean equals(DNA otherSeq) {
// TODO
return false;
}
/*
* This one should be easier: all too have to do here is print out
* the original DNA sequence. Make sure the String is either all
* upper case or all lower case (your choice, but I chose to work
* with all lower case).
*/
public String toString() {
// TODO
return null;
}
/*
* Return the complement DNA sequence of the original DNA sequence.
*/
public DNA toComplement() {
// TODO
return null;
}
/*
* This method should return whether a DNA sequence called otherSeq
* can be found within the original DNA sequence represented by
* this class. Of course, differences in upper vs. lower case should.
* be ignored. Return 1 if otherSeq is contained, otherwise return 0.
*/
public int contains(DNA otherSeq) {
// TODO
return 0;
}
/* Don't touch this method! */
public static void main(String[] args) {
/* Test the functionality of your different methods here. */
DNA seq1 = new DNA("TTCTcTCGCCAGcACTGTaATAGGCACTAaaAGAGTGAtG");
DNA seq2 = new DNA("ggcacta");
DNA seq3 = new DNA("GtGctGAGCTaagACGGCgt");
// Test length() method.
System.out.println("Testing length() method...");
System.out.print(seq1.length() + " ");
System.out.print(seq2.length());
System.out.println();
// Test getGCcontent() method.
System.out.println("Testing getGCcontent() method...");
System.out.print(seq1.getGCcontent() + " ");
System.out.print(seq3.getGCcontent());
System.out.println();
// Test equals() method.
System.out.println("Testing equals() method...");
System.out.print(seq1.equals(seq1) + " ");
System.out.print(seq2.equals(seq3));
System.out.println();
// Test toString() method.
System.out.println("Testing toString() method...");
System.out.print(seq2.toString() + " ");
System.out.print(seq3.toString());
System.out.println();
// Test toComplement() method.
System.out.println("Testing toComplement() method...");
System.out.print(seq2.toComplement().toString() + " ");
System.out.print(seq3.toComplement().toString());
System.out.println();
// Test contains() method.
System.out.println("Testing contains() method...");
System.out.print(seq1.contains(seq2) + " ");
System.out.print(seq2.contains(seq1));
System.out.println();
}
}
Note: In order to write the toComplement() method, please visit this link to understand what a DNA complement sequence is. The relevant snippet is included here:
The reverse complement of a DNA sequence is formed by reversing the letters, interchanging A and T and interchanging C and G. For example, the reverse complement of ACCTGAG is CTCAGGT.
This is the expected output so you can check your work:
$ javac DNA.java
$ java DNA
Testing length() method...
40 7
Testing getGCcontent() method...
0.45 0.6
Testing equals() method...
true false
Testing toString() method...
ggcacta gtgctgagctaagacggcgt
Testing toComplement() method...
tagtgcc acgccgtcttagctcagcac
Testing contains() method...
1 0
Problem 2
In this problem, we will investigate the differences ArrayLists and LinkedLists we talked about earlier. More specifically, LinkedLists are much faster at inserting and removing different list elements, while ArrayLists are much faster at accessing and writing to different list elements. First, create a new Java class called Compare.java, and copy-paste the following code.
import java.util.Calendar;
import java.util.*;
public class Compare {
private ArrayList<Integer> a;
private LinkedList<Integer> l;
public Compare(int numElements) {
this.a = new ArrayList<>(numElements);
this.l = new LinkedList<>();
for (int i = 0; i < numElements; i++) {
this.a.add(1);
this.l.add(1);
}
}
public long popArrayElement() {
long start = System.nanoTime();
this.a.remove(0);
long stop = System.nanoTime();
return (stop - start);
}
public long popLinkedElement() {
long start = System.nanoTime();
this.l.remove(0);
long stop = System.nanoTime();
return (stop - start);
}
public long accessArrayElement() {
long start = System.nanoTime();
int dummy = this.a.get(this.a.size() / 2);
long stop = System.nanoTime();
return (stop - start);
}
public long accessLinkedElement() {
long start = System.nanoTime();
int dummy = this.l.get(this.l.size() / 2);
long stop = System.nanoTime();
return (stop - start);
}
public void comparePop() {
long array = this.popArrayElement();
long linked = this.popLinkedElement();
System.out.println("It takes ArrayList " + array + " nsec to " +
"remove the first element in the list.");
System.out.println("It takes LinkedList " + linked + " nsec to " +
"remove the first element in the list.");
System.out.println("It takes ArrayList " + (array / linked) +
" times longer to remove the first element " +
"in the list.");
System.out.println();
}
public void compareAccess() {
long array = this.accessArrayElement();
long linked = this.accessLinkedElement();
System.out.println("It takes ArrayList " + array + " nsec to " +
"access an element in the middle of the list.");
System.out.println("It takes LinkedList " + linked + " nsec to " +
"access an element in the middle of the list.");
System.out.println("It takes LinkedList " + (linked / array) +
" times longer to access an element in the " +
"middle of the list.");
System.out.println();
}
public static void main(String[] args) {
Compare program = new Compare(Integer.parseInt(args[0]));
program.comparePop();
program.compareAccess();
}
}
You do not need to modify the code in any way. After you have done this, answer the following questions.
- Read the code and make sure you understand what the code is trying to accomplish. To learn more about the
System.nanoTime()method, check out this link. (Comment the code if it’s helpful!) - Run this program with 100 passed in as the argument. What do you notice?
- Run this program with 1000000 (one million) passed in as the argument. What do you notice?
- Run this program with 10000000 (ten million) passed in as the argument. What do you notice?
- Does your experiment results agree with your understanding of how
ArrayLists andLinkedLists work?
Make sure to repeat steps 2-4 a few times to make sure you get reproducible results. Your outputs will very likely not be the same between runs, but they should be within the same order of magnitude.